Sizhuang He
Loading...
Citations
Loading...
H-Index
Last updated: Loading...
About Me
I’m a second-year Computer Science Ph.D. student at Yale University, advised by Dr. David van Dijk. Previously, I completed my Bachelor’s degree at University of Michigan, Ann Arbor, majoring in Honors Mathematics and minoring in Computer Science.
I work on the intersection of machine learning and biology:
- Generative Modeling: Flow Matching, Diffusion, Discrete Diffusion
- Operator Learning: Modeling Continuous Spatiotemporal Dynamics, Integral Equations
- Computational Biology: Single-cell Transcriptomics Data Analysis
- LLMs and Agentic AI: Autonomous Systems for Biological Discovery
News
- [Sep 2025] Our paper Non-Markovian Discrete Diffusion with Causal Language Models is accepted to NeurIPS 2025 (San Diego, CA).
- [Jul 2025] Our paper COAST: Intelligent Time-Adaptive Neural Operators is accepted to AI4MATH@ICML 2025 (Vancouver, Canada).
- [Jan 2025] I received the Fan Family Fellowship of Yale University.
- [Jan 2025] Our paper Intelligence at the Edge of Chaos is accepted to ICLR 2025 (Singapore).
Selected Publications
-
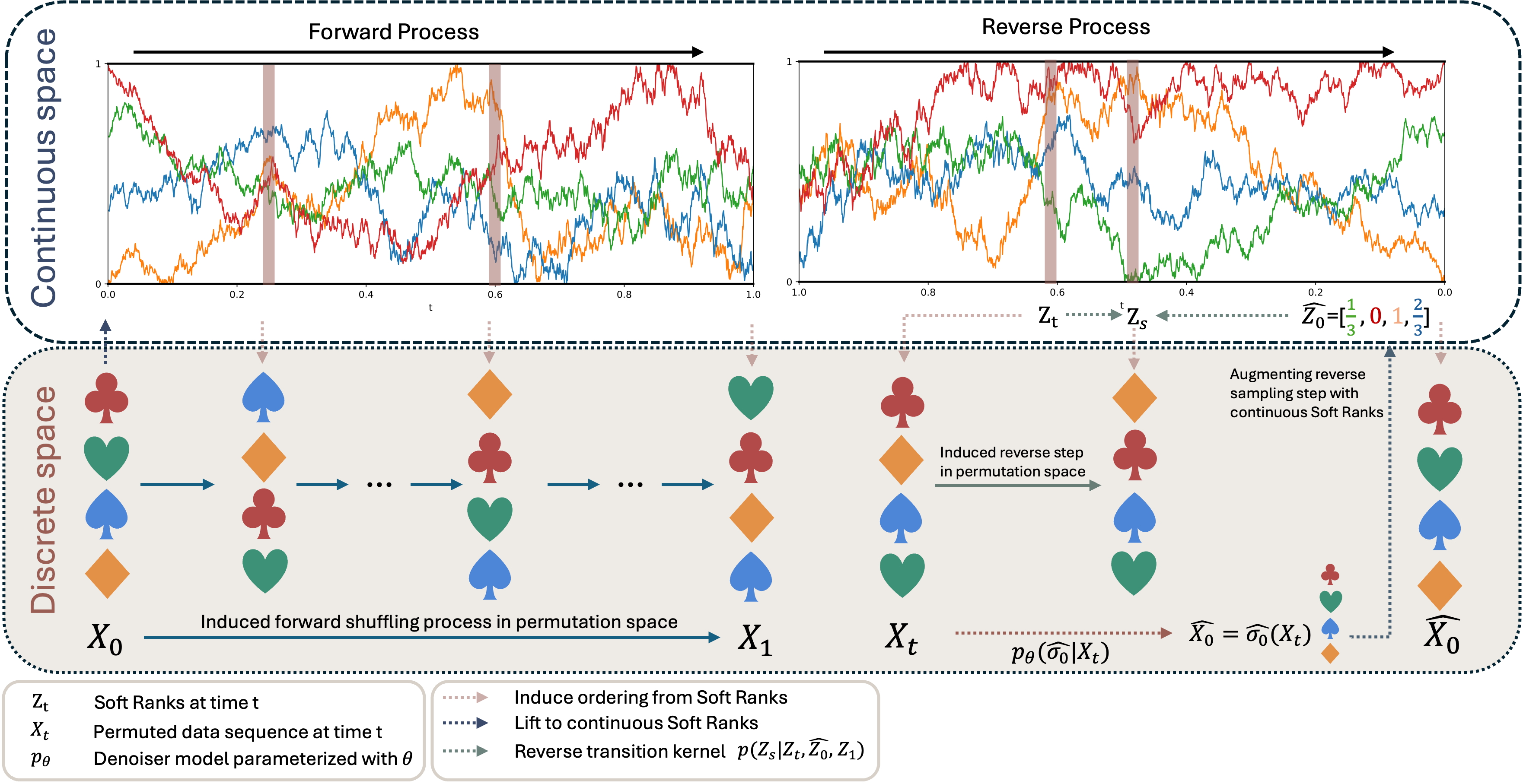 Soft-Rank DiffusionWe propose Soft-Rank Diffusion, a discrete diffusion framework for learning distributions over permutations on $S_n$ that replaces shuffle-based random walks with a structured soft-rank forward process. By lifting permutations to continuous relaxed ranks, we obtain smoother and more tractable trajectories, while our contextualized generalized Plackett–Luce (cGPL) denoisers provide a prefix-conditional generalization of PL-style reverse models for inherently sequential decision structure. Across sorting and combinatorial optimization benchmarks, Soft-Rank Diffusion consistently outperforms prior permutation diffusion baselines, with especially strong gains on long sequences. — In Review
Soft-Rank DiffusionWe propose Soft-Rank Diffusion, a discrete diffusion framework for learning distributions over permutations on $S_n$ that replaces shuffle-based random walks with a structured soft-rank forward process. By lifting permutations to continuous relaxed ranks, we obtain smoother and more tractable trajectories, while our contextualized generalized Plackett–Luce (cGPL) denoisers provide a prefix-conditional generalization of PL-style reverse models for inherently sequential decision structure. Across sorting and combinatorial optimization benchmarks, Soft-Rank Diffusion consistently outperforms prior permutation diffusion baselines, with especially strong gains on long sequences. — In Review -
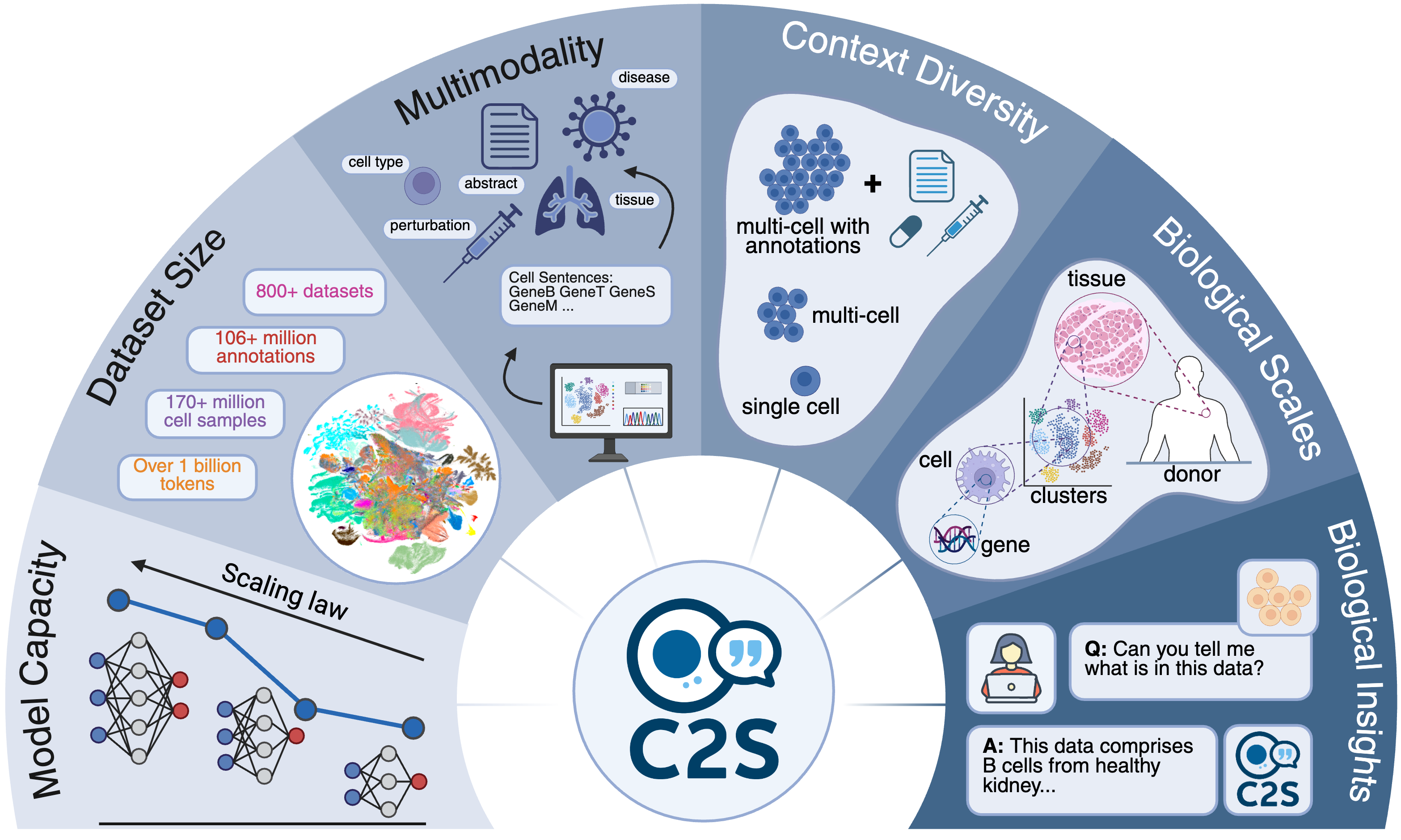
Cell2SentenceC2S-Scale scales this framework to 27 billion parameters trained on a billion-token multimodal corpus—achieving state-of-the-art predictive and generative performance for complex, multicellular analyses. Visit the project page. Read more about this work in our blog post and another blog post. — In Review
-
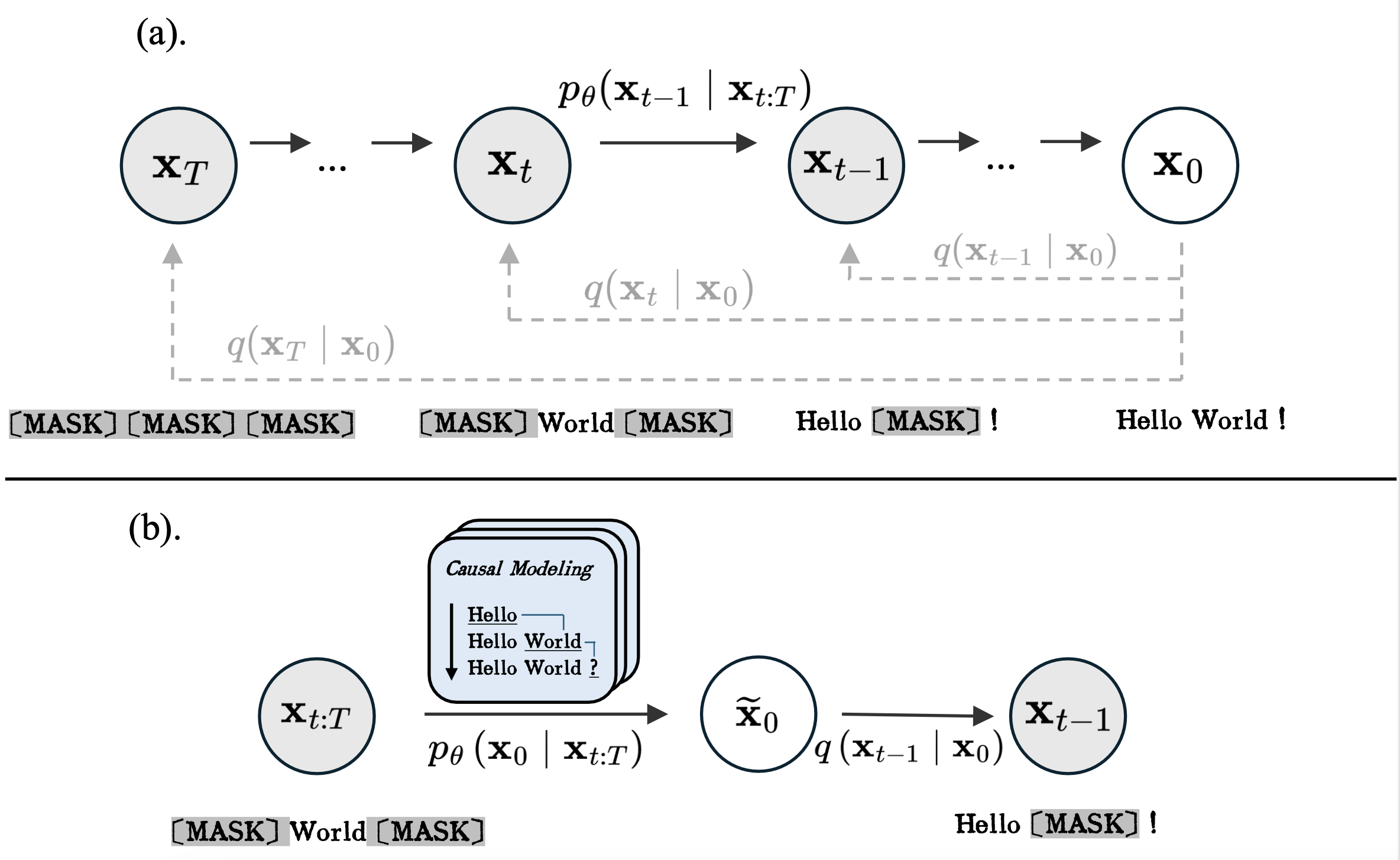 CaDDIWe introduce a novel approach to discrete diffusion models that conditions on the entire generative trajectory, thereby lifting the Markov constraint and allowing the model to revisit and improve past states. CaDDi treats standard causal language models as a special case and permits the direct reuse of pretrained LLM weights with no architectural changes. — NeurIPS 2025
CaDDIWe introduce a novel approach to discrete diffusion models that conditions on the entire generative trajectory, thereby lifting the Markov constraint and allowing the model to revisit and improve past states. CaDDi treats standard causal language models as a special case and permits the direct reuse of pretrained LLM weights with no architectural changes. — NeurIPS 2025 -
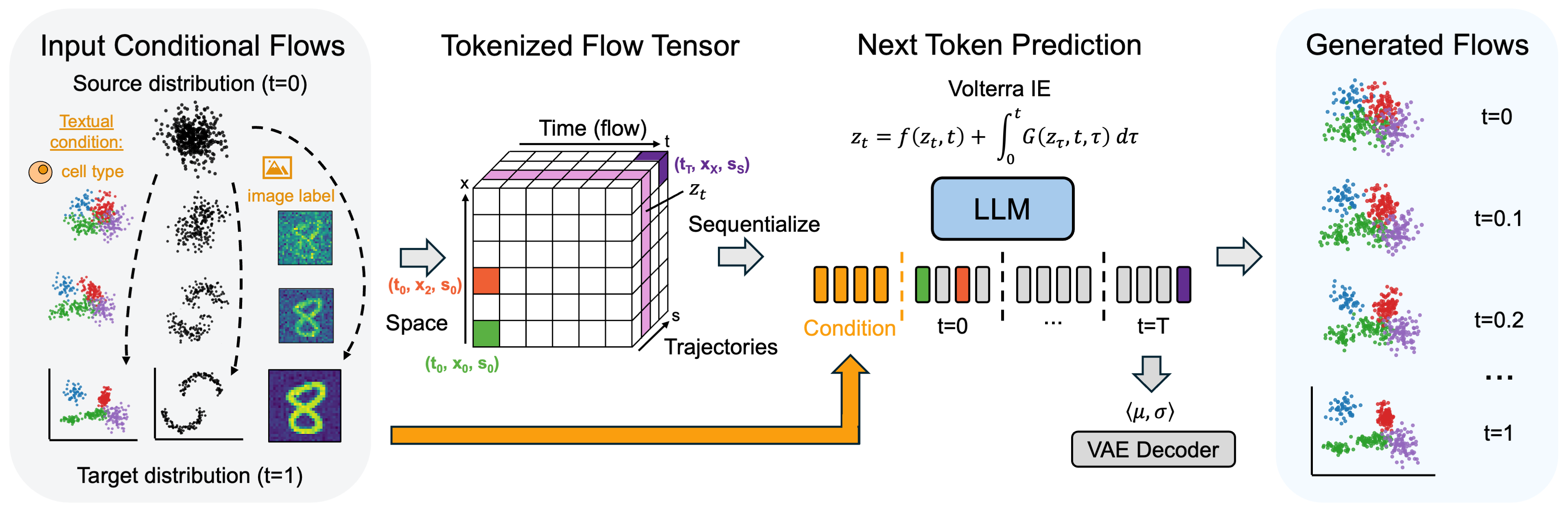
CaLMFlowWe present Volterra Flow Matching, a novel generative modeling framework that reformulates ODE-based flow matching frameworks with Volterra Integral Equations, hence avoiding a core challenge in ODE-based methods, known as stiffness. We show the connection between Volterra Integral Equations and causal transformers, the backbone of modern Large Language Models and hence demonstrates that causal language models can be naturally extended to generative modeling over continuous data domains through the lens of Volterra Flow Matching. — arXiv
-
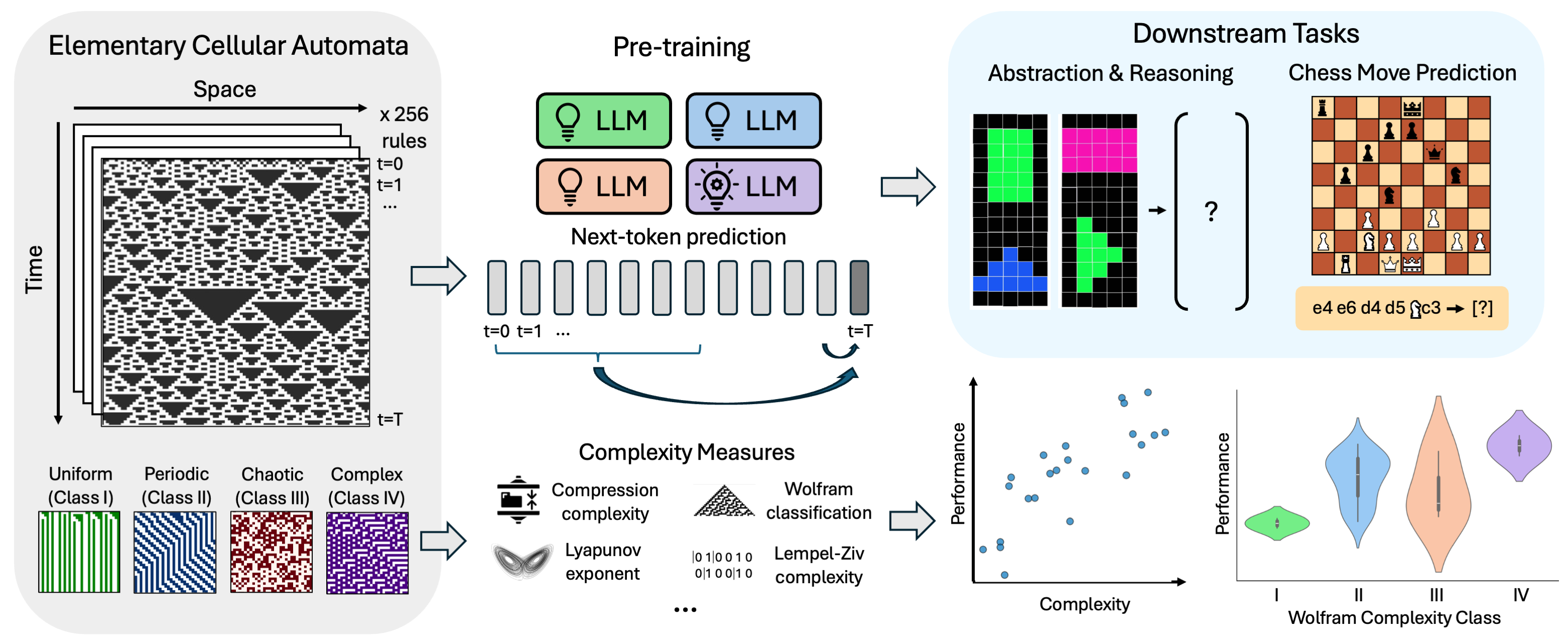 Intelligence at the Edge of ChaosBy training LLMs on elementary cellular automata rules of varying complexity, we pinpoint a ‘sweet spot’ of data complexity that maximizes downstream predictive and reasoning abilities. Our findings suggest that exposing models to appropriately complex patterns is key to unlocking emergent intelligence. — ICLR 2025
Intelligence at the Edge of ChaosBy training LLMs on elementary cellular automata rules of varying complexity, we pinpoint a ‘sweet spot’ of data complexity that maximizes downstream predictive and reasoning abilities. Our findings suggest that exposing models to appropriately complex patterns is key to unlocking emergent intelligence. — ICLR 2025
Services
Journal Reviewer
- ACM Computing Surveys
- Transactions on Machine Learning Research
Conference Reviewer
- International Conference on Machine Learning, 2026
- International Conference on Learning Representations, 2026
- AI4MATH Workshop at International Conference on Machine Learning, 2025
Powered by Jekyll and Minimal Light theme.